IGV frontend (desktop client) setup
Integrative Genomics Viewer for AWS Cloud
This blogpost assumes assumes that you have your AWS infrastrucure already in place, if not, read the Amazon IGV backend setup.
At UMCCR, we use AWS to access the BAM and VCF files. Other ways of accessing data are discouraged for security and practical reasons… unless you use IGV.js or other browser-based systems, which are preferred.
Use Amazon
- If you had previous manually-crafted
oauth-config.jsonfile(s) under your IGV user directory, please remove it. - If you are on a Mac, install with Homebrew with
brew cask install igv. Alternatively, manually download from the official Broad site: https://software.broadinstitute.org/software/igv/download and download only versions >=2.7.2, older releases will not work. - Use the following OAUTH provisioning URL in your IGV advanced preferences (
View->Preferences->Advanced->"OAuth provisioning URL"), namely:
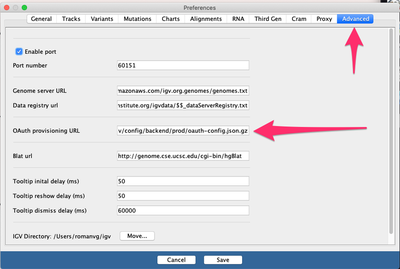
The URL to copy and paste on the box above is this: https://raw.githubusercontent.com/umccr/infrastructure/master/cdk/apps/igv/config/backend/prod/oauth-config.json.gz
To operate the Amazon features, the easiest way is to use the UMCCR Data Portal… if you work at UMCCR:
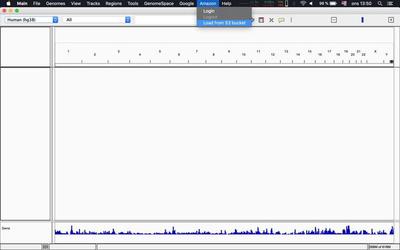
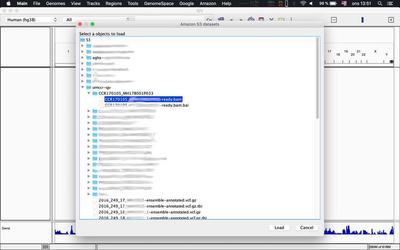
Alternatively you can load tracks as described in this tweet from within IGV itself:
Use dev environment (optional)
NOTE: This feature will only work with IGV desktop versions 2.8.9 and above
If you are an advanced user that needs to have two concurrent IGV installations running on the same computer, connected to development and production environments, you can create separate IGV user directories that contain special configuration:
Provision your IGV instance as described above with the following URL:
Known issues/FAQ
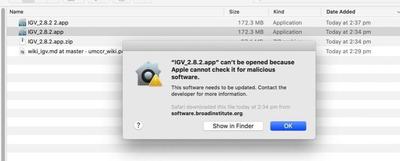
Q: When I try to install IGV I’m faced with a "… can’t be opened because Apple cannot check it for malicious software", such as:
Q: I cannot see the Amazon menu on my IGV, what’s wrong?
A: Please revise the “Provisioning URL” copy and pasted from above (or from a PDF) very carefully and make sure it does NOT contain spaces.
A: This is a known issue with OSX Catalina, see what the IGV team has to say about it: https://github.com/igvteam/igv/issues/713#issuecomment-542362787
Q: IGV is really sluggish, fails to load data and/or says “Out of memory, stopping read reading” or similar issues.
A: Please adjust the default -Xmx4g memory setting to -Xmx8g or higher, depending on your system’s available memory.
Q: I am a PeterMac employee and I’m using its network. Will I be able to use IGV with AWS?
A: No. The PMCC network apparently blocks IGV requests at an application level, not much we can do about it.
Q: When I try to install IGV I’m faced with a “This app is Damaged” error.
A: Third party applications installed by “unkwnown developers” (not signed apps) are probably not allowed try running sudo spctl --master-disable, run IGV and then sudo spctl --master-enable after it’s up and running.
Q: On windows, the UMMCR-IGV app suddenly opens a terminal and closes it immediately, nothing happens.
A: Most probably the available RAM memory on your computer is too little to allocate Java’s heap space. Please locate and edit the igv.bat file and modify the parameter Xmx4g to, for instance, Xmx1g if you happen to only have 2GB of RAM (older computers).
Q: When I enter a coordinate on the search box, the progress spinner waits indefinitely, nothing loads, cannot see any reads from the BAM file I loaded.
A: IGV is picky with chromosome coordinates, please specify those without commas, but with a big integer instead, i.e: chr1:100043 instead of chr1:100,000. Other than that, there’s a know issue about that behaviour: it generally means that the region you are exploring is just N’s, this only happens on windows.
Q: ERROR [2019-09-26T11:21:33,975] [CommandListener.java:140] [Thread-3] java.net.BindException: Address already in use: NET_Bind
A: Do you have another IGV process/session running in parallel? Close it. Still not working? Reboot your computer.
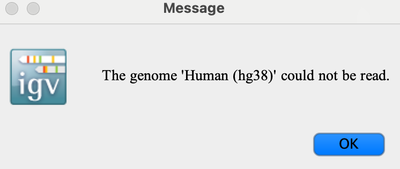
Q: “The genome ‘Human (hg38) could not be read”
$HOME and configure your organization’s OAuth access as outlined in the beginning of this blogpost.