AWS Rust Lambdas and Bioinformatics
Yet another AWS Lambda and Rust blogpost?
At this point in time, much has been written about AWS and Rust Lambdas, here’s a personal selection in no particular order: 0 1 2 3 4 5 6 7
This writeup patches most of those blogposts with a couple of recent developments and proposes a cool (paid) project to work on if the reader is keen:
- The latest, first officially released on crates.io, aws-lambda-rust-runtime version 0.3.0 and why you don’t need MUSL in your lambda builds anymore. If you are interested in quickly adopting those changes, read the “New lambda runtime” section below.
- Proposes an interesting BioIT project for students, that uses the tech described in this article. This is paid by Google if you are accepted on this year’s GSoC htsget-rs idea under the GA4GH organization.
- On the bioinformatics front, we outline a serialization-deserialization (BioSerDe?) future where file formats get abstracted to allow easier experimentation with different storage and compute backends.
So, without further ado, what’s new in the AWS Rust serverless space?
New AWS lambda runtime
Released just a few weeks ago by a relentless effort from (mostly, among others) @bahildebrand and @coltonweaver, release 0.3.0 most likely breaks some of the examples from the blogposts listed above in the introduction.
While the official documentation for this crate was updated accordingly, you might want to clone s3-rust-noodles-bam as a testbed, since it includes the improvements described in this blogpost and the application’s payload is hopefully more interesting than a hello world.
Letting go of MUSL static builds
One of the ugly truths of AWS lambdas is that the underlying “provided” Amazon Linux v1 build is too old to link against more recent GLIBCs, often yielding errors like:
(...)
/var/task/bootstrap: /lib64/libc.so.6: version `GLIBC_2.18' not found (required by /var/task/bootstrap)
(...)
ms Billed Duration: 100 ms Memory Size: 128 MB Max Memory Used: 12 MB
Until the AWS lambda provided.al2 was released, the workaround was to build the resulting bootstrap (lambda payload binary) with MUSL. This means statically linking your payload and all dependencies, inflating the resulting binary and generating many headaches on the integration and deployment land.
No more.
Just use provided.al2 and target x86_64-unknown-linux-gnu and jemallocator. The latter just requires a couple of simple changes on your Cargo.toml and main.rs, refer to the docs.
So now we have dynamically linked GLIBC for x86_64 with an optimized memory allocator. That’s at least until AWS lambdas don’t run on Graviton2 (arm64)? Also, speaking of Lambda deployment…
Embrace AWS SAM…
… while AWS fixes its reported Rust-specific developer experience issues.
Until then, TMTOWTDI for AWS Rust Lambdas: Manually upload the resulting zipped bootstrap, Terraform, CDK, serverless.com and AWS’s SAM, so pick and choose.
After testing a few of those approaches on s3-rust-htslib-bam, we’ll be sticking with SAM since its tooling is meant to integrate well with AWS.
Having the updates and gotchas covered, let’s talk about how to use this great tech next?
Motivation
Shortly after announcing that this project idea was up for grabs, a few voices pointed out some good questions:
There is already a Golang implementation of htsget IIRC, and so one for C4GH: So building this in go could be easier, although perhaps does not live up to performance expectations?
I would like to know the reasons for choosing rust for this task primarily. I am a fan of the language but given what’s out there now it feels a bit of an overkill?
Here’s a few reasons we think that justify the time and effort to implement this project:
- Multiple independent implementations help establish standards and allow us to demonstrate interoperability; it also shows where standards/references are ambiguous or underspecified.
- While optimizing the implemention of this project we will most probably also help related Rust (crates), nourishing its ecosystem as a result. Diverse software options are important and sorely needed in our field, htslib has been central to Bioinformatics for many years now and htsjdk has its own issues too.
- UMCCR’s @victorskl pull-requested an Amazon-specific AWS-Go-SDK integration for the GA4GH’s Go implementation, which is currently under scrutiny. Perhaps that pullrequest will never be merged to stay on principle: a reference implementation should be platform-agnostic. On the other hand, this Rust project is highly opinionated and does not have that particular limitation.
- The reference implementation just shells out to
samtoolsunder the hood. This means that it still useshtslib, with integration issues such as the one noted above. - Safety, see comments about OpenSSL and htslib. many static analysis via fuzzers reveal memory leaks and other defects found routinely on htslib this way. A pure Rust implementation does not have to go through those audits because the compiler guarantees a series of safety measures against those defects.
- Performance vs Go argument? See a well written, reality-based and pragmatic justification here: https://thenewstack.io/rust-vs-go-why-theyre-better-together/?s=09
To be fair, regarding performance, there’s both Fargate and Lambda supporting docker images now, which probably provides a more than good enough solution?
Since towards the end of this project we will run some benchmarks and see what makes sense to use in different regimes.
Google Summer of Code 2021 and htsget-rs
The GA4GH (Genomics Alliance for Genomics and Health) was accepted as an organization for this year’s (GSoC) Google’s Summer of Code.
Expectations
Since Rust libraries in our field are currently developing, this project has a strong community component. In our proposal we will inevitably be touching a few crates from the Rust ecosystem:
- Noodles
- AWS Rust Lambda Runtime
- Rust-S3 (as a temporal replacement to Rusoto).
- RustLS (as a permanent replacement to OpenSSL).
Unexpected issues might crop up on the AWS side or elsewhere. This means that the student must be prepared to fix third party dependencies and thus interact with different crate authors and communities, with some help from the mentors if need be.
There’s also some minor onboarding particularities for this project, namely having access to an Amazon Web Services account: Our team at UMCCR can provide temporal access to a limited AWS account for this particular project. In addition, the successful student will be added to GA4GH’s #htsget Slack channel as well as pointers to the broader Rust community.
Architecture and implementation plan
As outlined in the official htsget specification, only two API endpoints are required: reads and variants (we are obviating ticket handling for simplicity in this writeup):
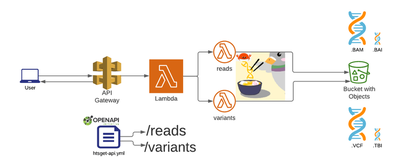
GA4GH already has a reference htsget implementation written in Go, partially based on an previous older Google-specific implementation.
This presents itself as a huge advantage since we can examine parts of it like the endpoints defined by the ga4gh-refserver implementation and build the aforementioned mock testing outlined above. Many other functional groups that conform a htsget implementation are already very well laid out:
That being said, Rust not only has different syntax, module structure and other primitives than Go, but it also has its own supported ways to structure projects via Cargo workspaces.
For instance, Noodles itself uses cargo workspaces to modularise and compose different file format structures and their associated functions:
[workspace]
members = [
"noodles",
"noodles-bam",
(...)
"noodles-sam",
"noodles-tabix",
"noodles-vcf",
]
Furthermore, since Michael had in mind a Rust implementation of htsget when he started Noodles, there are functions worth keeping in mind that will prove useful while implementing the low level details of parsing a BAI file and optimizing byte ranges, see optimize_chunks. This detail will come handy later on during the implementation, but it’s not important right now, so don’t worry if it’s not clear at the moment.
When a local implementation works it’ll be time to leverage the work already done on the newest Noodles+Rust-S3+AWS+Lambda example. Once this stage is reached, it is important to remember the system description graphic above and realise that the scope for our implementation deals with S3 public buckets since all the authZ/authN happens on the AWS API Gateway. In other words, we will not be writing code that validates JWT tokens ourselves since that is already managed by AWS’s API Gateway.
The Go htsget reference implementation decouples security from core application functionality. In this implementation, we’ll also do that but assume that data is fronted by a managed service such as API Gateway.
Last but not least, benchmarking or “benches” needs to be run to assess bottlenecks and aim at future improvements iteratively.
Ready to start writing some Rust with us for GA4GH’s GSoC? Jump to the next section!
Qualifying task
Implementing the following task, while not mandatory, gives extra points to candidates since it helps mentors with the student selection process.
The first step the student should take is implementing a local (no AWS involved) htsget interface. This step is important to accomplish while we review proposals since it gives mentors and indication of:
- How resourceful the incumbent is when carrying on API development.
- Which code structure is followed to attain this goal? Do they constitute good use of abstractions, data structures and overall code quality?
- Which questions are being asked to mentors and how thoughtful, precise are those?
- Does the student manage version control and collaborative development successfully?
The task would be to understand, code and pullrequest a substantial constribution to this skeleton repo:
https://github.com/chris-zen/htsget-mvp
Good luck and see you soon! :)
Future
TL;DR: EVERYTHING IS SERIALIZATION
This section is out of scope for GSoC, but outlines a few exciting followup experiments that could materialize in the future:
- Generalize Robert’s Aboukhalil WASM+CloudFlare experiment. Use htsget as comms layer?
- Think forward about (de)serializing our niche bioinformatics formats to other supported formats by the Rust SerDe crate. There is an ongoing approach by Mike Linn SQLite Genomics which is a particular case of a more general model, as in BAM to: Arrow, Presto UDFs or any destination format used by data analysis frameworks outside bioinformatics.
- Use those serializers mentioned above in other tooling such as more genome visualization apps or even into Radare2 so that other communitites and fields can easily interact with genomics data.
- Interface with the ADAM ecosystem, perhaps combining with vega, formerly known as native-spark.
Thanks to @chris-zen, @victorskl, @zaeleus and the UMCCR team for proofreading and suggesting improvements for this blogpost.